Learn more: PMC Disclaimer | PMC Copyright Notice
The genome sequence of a 45,000-year-old modern human from western Siberia
Associated Data
Abstract
We present a high-quality genome sequence of a ~45,000-year-old modern human male from Siberia. This individual derives from a population that lived prior to – or simultaneously with – the separation of the populations in western and eastern Eurasia and carries a similar amount of Neandertal ancestry as present-day Eurasians. However, the genomic segments of Neandertal ancestry are substantially longer than those observed in present-day individuals, indicating that Neandertal gene flow into the ancestors of this individual occurred 7,000–13,000 years before he lived. We estimate an autosomal mutation rate of 0.4–0.6×10−9/site/year and a Y chromosomal mutation rate of 0.7–0.9×10−9/site/year based on the additional substitutions that have occurred in present-day non-Africans compared to this genome, and a mitochondrial mutation rate of 1.8–3.2 × 10−8/site/year based on the age of the bone.
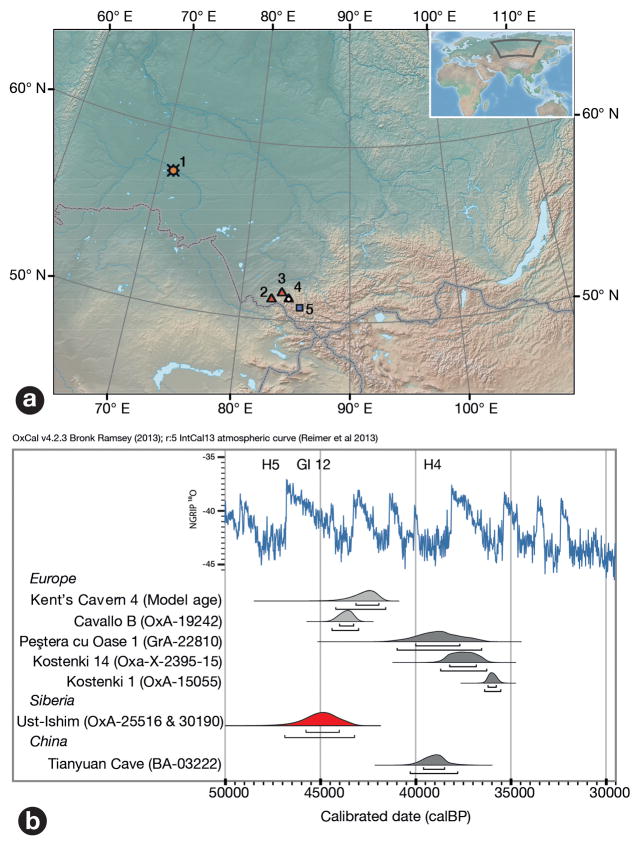
In 2008, a relatively complete left human femoral diaphysis was discovered on the banks of the river Irtysh (Fig. 1a, c, d), near the settlement of Ust’-Ishim in western Siberia (Omsk Oblast, Russian Federation). Although the exact locality is unclear, the femur was eroding out of alluvial deposits on the left bank of the river, north of Ust’-Ishim. Here, Late Pleistocene and probably redeposited Middle Pleistocene fossils are found in sand and gravel layers that are about 50,000–30,000 years old (i.e. from Marine Oxygen Isotope Stage 3).
a) Map of Siberia with major archaeological sites. Triangles: Neandertal fossils; white circle in triangle: Denisovan fossils; blue squares: Initial Upper Palaeolithic sites; orange symbol: Ust’-Ishim. 1: Ust’-Ishim; 2: Chagyrskaya Cave; 3: Okladnikov Cave; 4: Denisova Cave; 5: Kara-Bom; B) Radiocarbon ages of early modern human fossils in Northern Eurasia and the NGRIP δ18O palaeotemperature record. Specimens in light grey are indirectly dated. H5: Heinrich 5 event, H4: Heinrich 4 event, GI 12: Greenland Interstadial 12. For a more extensive comparison see Figure SI2.1. c–f) The Ust’-Ishim 1 femur. c) lateral view; d) posterior view; e) cross section at the 80 percent level; f) cross section at the midshaft. For other views see Figure S3.1.
The proximal end of the bone shows a large gluteal buttress and gluteal tuberosity, while the midshaft is dominated by a marked Linea aspera, resulting in a teardrop-shaped cross section (Fig. 1, e,f) (for details, see SI 3). The morphology of the proximal end of the shaft is similar to Upper Paleolithic modern humans and distinct from Neandertals (Table S3.1, Fig. S3.2.) while the teardrop-shaped cross section of the midshaft is similar to most Upper Paleolithic humans and early anatomically modern humans 1. Taken together, this suggests that the Ust’-Ishim femur derives from a modern human.
Two samples of 890 mg and 450 mg of the bone was removed on separate occasions for dating. Collagen preservation satisfied all criteria for dating 2 and after ultrafiltration we obtained ages of 41,400 ± 1300 years before present (yrs BP) (OxA-25516) and 41,400 ± 1400 BP (OxA-30190). These two dates, when combined and corrected for fluctuations of atmospheric 14C through time correspond to an age of about 45,000 calibrated years BP (46,880–43,210 cal BP at 95.4% probability, SI 1) The Ust’-Ishim individual is therefore the oldest directly radiocarbon-dated modern human outside Africa and the Middle East (Fig. 1B). Carbon and nitrogen isotope ratios indicate that the diet of the Ust’-Ishim individual (SI 4) was based on terrestrial C3 plants and animals that consumed them, and that an important part of his dietary protein may have come from aquatic foods, probably freshwater fish, something that has been observed in other early Upper Palaeolithic humans from Europe 3.
Nine samples of between 41 and 130 mg of bone material were removed from the distal part of the femur and used to construct DNA libraries using a protocol designed to facilitate the retrieval of short and damaged DNA 4. The percent of DNA fragments in these libraries that could be mapped to the human genome varied between 1.8% and 10.0% (Table S1.1). From the extract containing the highest proportion of human DNA, eight further libraries were constructed. Each of these libraries was treated with uracil-DNA-glycosylase and endonuclease VIII to remove deaminated cytosine residues, and library molecules with inserts shorter than approximately 35 base pairs (bp) were depleted by preparative acrylamide gel electrophoresis prior to sequencing on the Illumina HiSeq platform (SI 6). In total, 42-fold sequence coverage of the ~1.86 gigabases (Gb) of the autosomal genome to which short fragments can be confidently mapped was generated. The coverage of the X- and Y-chromosomes was approximately half that of the autosomes (~22-fold), indicating that the bone comes from a male. A likelihood method estimated present-day human mitochondrial DNA (mtDNA) contamination 5 to 0.50% (95% confidence interval (C.I.) 0.26%–0.94%) while a method that uses the frequency of non-consensus bases in autosomal sequences estimated the contamination to be less than 0.13% (SI 7). Thus, less than 1% of the hominin DNA fragments sequenced are estimated to be extraneous to the bone. After consensus genotype calling such low levels of contamination will tend to be eliminated.
About 7.7 positions per 10,000 are heterozygous in the Ust’-Ishim genome while between 9.6 and 10.5 positions are heterozygous in present-day Africans and 5.5 and 7.7 in present-day non-Africans (SI 12). Thus, with respect to genetic diversity, the population to which the Ust’-Ishim individual belonged was more similar to present-day Eurasians than to present-day Africans, which likely reflects the out-of-Africa bottleneck shared by non-African populations. The Ust’-Ishim mtDNA sequence falls at the root of a large group of related mtDNAs (the “R haplogroup”), which occurs today across Eurasia (SI 8). The Y chromosome sequence of the Ust’-Ishim individual is similarly inferred to be ancestral to a group of related Y chromosomes (haplogroup K(xLT)) that occurs across Eurasia today 6 (SI 9). As expected, the number of mutations inferred to have occurred on the branch leading to the Ust’-Ishim mtDNA is lower than the numbers inferred to have occurred on the branches leading to related present-day mtDNAs (Fig. S8.1). Using this observation and nine directly carbon-dated ancient modern human mtDNAs as calibration points 5,7 in a relaxed molecular clock model, we estimate the age of the Ust’-Ishim bone to be ~49,000 yrs BP (95% highest posterior density: 31,000–66,000 yrs BP), consistent with the radiocarbon date.
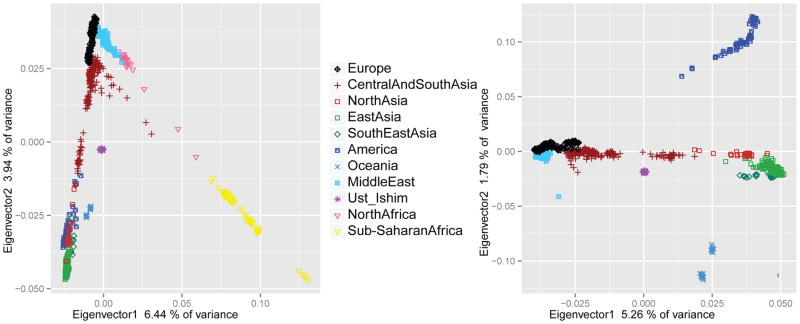
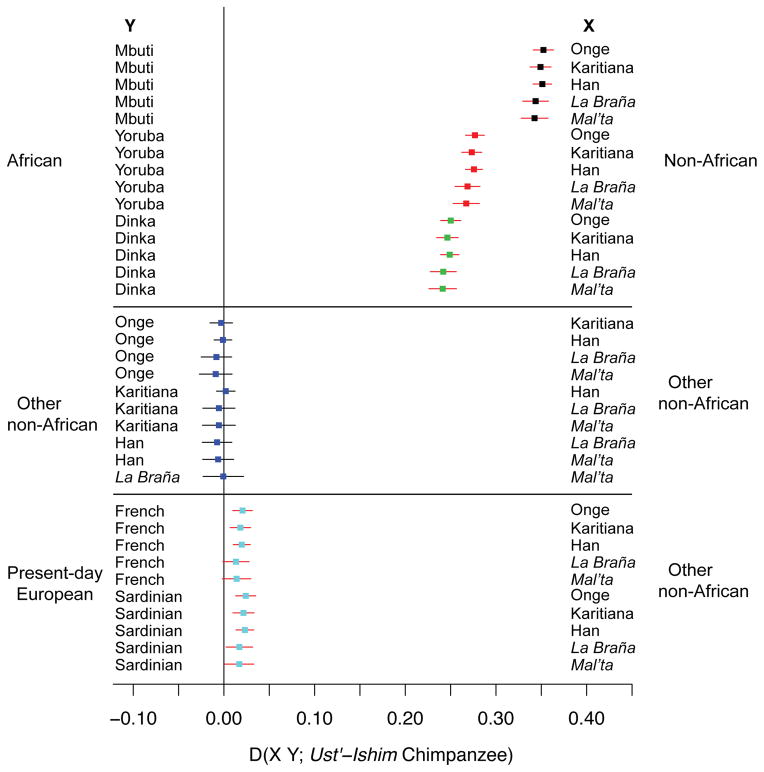
In a principal component (PC) analysis of the Ust’-Ishim autosomal genome along with genotyping data from 922 present-day individuals from 53 populations 8 (Fig. 2A), the Ust’-Ishim individual clusters with non-Africans rather than Africans. When only non-African populations are analyzed (Fig. 2B), the Ust’-Ishim individual falls close to zero on the two first PC axes suggesting that it does not share much more ancestry with any particular group of present-day humans than with any other. To determine how the Ust’-Ishim genome is related to the genomes of present-day humans, we tested, using “D-statistics” 8, whether it shares more derived alleles with one modern human than with another using pairs of human genomes from different parts of the world (Fig. 3). Based on genotyping data for 87 African and 108 non-African individuals (SI 11), the Ust’-Ishim genome shares more alleles with non-Africans than with sub-Saharan Africans (|Z| = 41–89), consistent with the PCA, mtDNA and Y chromosome results. Thus, the Ust’-Ishim individual represents a population derived from, or related to, the population involved in the dispersal of modern humans out of Africa. Among the non-Africans, the Ust’-Ishim genome shares more derived alleles with present-day people from East Asia than with present-day Europeans (|Z| = 2.1–6.4). However, when an ~8,000-year-old genome from western Europe (La Braña) 9 or a 24,000-year-old genome from Siberia (Mal’ta 1) 10 were analyzed, there is no evidence that the Ust’-Ishim genome shares more derived alleles with present-day East Asians than with these prehistoric individuals (|Z| < 2). This suggests that the population to which the Ust’-Ishim individual belonged diverged from the ancestors of present-day West Eurasian and East Eurasian populations prior to - or simultaneously with - their divergence from each other. The finding that the Ust’-Ishim individual is equally closely related to present-day Asians and to 8,000- to 24,000-year-old individuals from western Eurasia, but not to present-day Europeans, is compatible with the hypothesis that present-day Europeans derive some of their ancestry from a population that did not participate in the initial dispersals of modern humans into Europe and Asia 11.
A) Principal Components (PC) analysis using 922 present-day individuals from 53 populations and the Ust’-Ishim individual; B) PC analysis using Eurasian individuals and the Ust’-Ishim individual. The percentages of the total variance explained by each eigenvector are given.
Statistics testing whether the Ust’-Ishim genome shares more derived alleles with one or the other of two modern human genomes (X, Y). We compute statistics of the form D (X, Y, Ust’-Ishim, Chimpanzee) using a subset of the genome-wide SNP array data from the Affymetrix Human Origins array and restricting the analysis to transversions. Error bars correspond to three standard errors. Red indicates that the D-statistic is significantly different from 0 (|Z|>2), such that the Ust’-Ishim genome shares more derived alleles with the genome on the right (X) than the left (Y). Ancient genomes are given in italics.
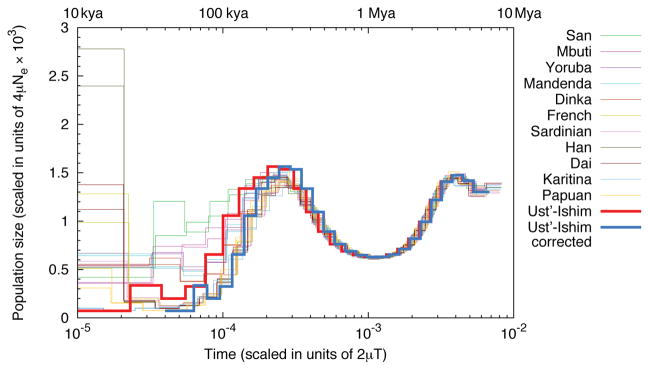
The high-quality Ust’-Ishim genome sequence, in combination with its radiocarbon date, allows us to gauge the rate of mutations by estimating the numbers of mutations that are “missing” in the Ust’-Ishim individual relative to present-day humans. This results in a mutation rate estimate of 0.44–0.63 × 10−9 using the high-coverage genomes of 14 present-day humans. A challenge for inferring the mutation rate in this way is that differences in error rates among genome sequences can confound the inference (see discussion in 12). We therefore developed an alternative approach that leverages the Pairwise Sequentially Markovian Coalescent (PSMC), a method which estimates the distribution of coalescence times between the two chromosomes across a diploid genome to estimate past changes in population size 13, and which is less influenced by differences in error rates. When the Ust’-Ishim genome along with 25 present-day human genomes are analyzed by PSMC, a recent reduction in population size similar to that seen for 11 present-day non-Africans is inferred for the Ust’-Ishim genome. However, the apparent age of this size reduction is more recent than in present-day humans, consistent with the Ust’-Ishim genome being older (Fig. 4). We then compute the number of additional substitutions that are needed to best fit the Ust’-Ishim PSMC curve to those of other non-African genomes. Assuming that this corresponds to the number of mutations that have accumulated over around 45,000 years, we estimate a mutation rate of 0.43 (95% C.I. 0.38 – 0.49 × 10−9/site/year) that is consistent across all non-African genomes regardless of their coverage (SI 14). This overall rate, as well as the relative rates inferred for different mutational classes (transversions, non-CpG transitions, and CpG transitions), is similar to the rate observed for de novo estimates from human pedigrees (~0.5 x10−9/site/year 14,15) and to the direct estimate of branch shortening (SI 10). As discussed elsewhere 14,16,17, these rates are slower than those estimated using calibrations based on the fossil record and thus suggest older dates for the splits of modern human and archaic populations. We caution, however, that rates may have changed over time and may differ between human populations. However, we expect this mutation rate estimate to apply at least to non-African populations over the past 45,000 years.
Inferred population size changes over time. “Time” on the X axis refers to the pairwise per-site sequence divergence. If we erroneously assume that Ust’-Ishim lived today, its inferred population size history includes an out-of-Africa-like population bottleneck that is more recent than that seen in present-day non-Africans (red bold curve). By shifting the Ust’-Ishim curve to align with those in present-day non-Africans (blue bold curve), and assuming that the number of mutations necessary to do this corresponds to 45,000 years, we estimate the autosomal mutation rate to 0.38–0.49 × 10−9/site/year. The times indicated on the top of the figures, are based on this mutation rate.
We also estimated a phylogeny relating the non-recombining part of the Ust’-Ishim Y chromosome to those of 23 present-day males. Using this phylogeny, we measured the number of “missing” mutations in the Ust’-Ishim Y chromosomal lineage relative to the most closely related present-day Y chromosome analyzed. This results in an estimate of the Y chromosome mutation rate of 0.76 × 10−9/site/year (95% C.I. 0.67–0.86 × 10−9/site/year) (SI 9), significantly higher than the autosomal mutation rate, consistent with mutation rates in males being higher than in females 18–20. Finally, using the radiocarbon date of the Ust’-Ishim femur together with the mtDNAs of 311 present-day humans, we estimated the mutation rate of the complete mtDNA to 2.53 × 10−8 substitutions per site per year (95% highest posterior density: 1.76–3.23 × 10−8) (SI 8) for mtDNA, in agreement with a previous study 5.
The time of admixture between modern humans and Neandertals has previously been estimated to 37,000–86,000 years BP based on the size of the DNA segments contributed by Neandertals to present-day non-Africans 21. Thus, the Ust’-Ishim individual could predate the Neandertal admixture. From the extent of sharing of derived alleles between the Neandertal and the Ust’-Ishim genomes we estimate the proportion of Neandertal admixture in the Ust’-Ishim individual to 2.3±0.3% (SI 16), similar to present-day East Asians (1.7–2.1%) and present-day Europeans (1.6–1.8%). Thus, admixture with Neandertals had already occurred by 45,000 years ago. In contrast, we fail to detect any contribution from Denisovans although such a contribution exists in present-day people not only in Oceania 22,23 but to a lesser extent also in mainland East Asia 12,24 (SI 17).
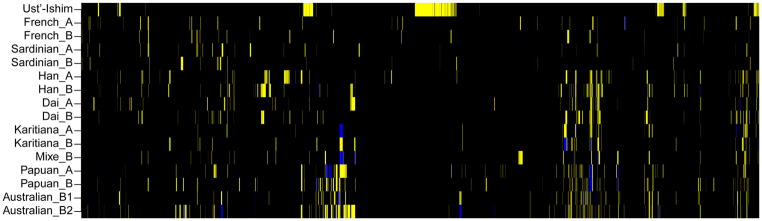
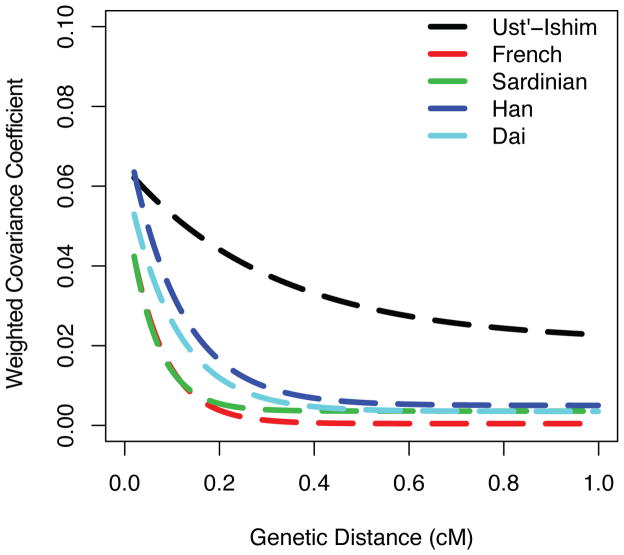
The DNA segments contributed by Neandertals to the Ust’-Ishim individual are expected to be longer than such segments in present-day people since the Ust’-Ishim individual lived closer in time to when the admixture occurred so that there was less time for the segments to be fragmented by recombination. To test if this is indeed the case, we identified putative Neandertal DNA segments in the Ust’-Ishim and present-day genomes based on derived alleles shared with the Neandertal genome at positions where Africans are fixed for ancestral alleles. Fig. 5 shows that segments of putative Neandertal origin in the Ust’-Ishim individual are substantially longer than those in present-day humans. We use the covariance in such derived alleles of putative Neandertal origin across the Ust’-Ishim genome to infer that mean fragment sizes in the Ust’-Ishim genome are in the order of ~1.8–4.2 times longer than in present-day genomes and that the Neandertal gene flow occurred 232–430 generations before the Ust’-Ishim individual lived (SI 18; Fig. 6). Under the simplifying assumption that the gene flow occurred as a single event, and assuming a generation time of 29 years 16,25, we estimate that the admixture between the ancestors of the Ust’-Ishim individual and Neandertals occurred approximately 50,000 to 60,000 years BP which is close to the time of the major expansion of modern humans out of Africa and the Middle East. This suggests that the bulk of the Neandertal contribution to present-day people outside Africa does not go back to mixture between Neandertals and the anatomically modern humans who lived in the Middle East at earlier times; for example, the modern humans whose remains have been found at Skhul and Qafzeh 26,27.
Regions of Neandertal ancestry on chromosome 12 in the Ust’-Ishim individual and fifteen present-day humans. The analysis is based on SNPs where African genomes carry the ancestral allele and the Neandertal genome carries the derived allele. Homozygous ancestral alleles are black, heterozygous derived alleles yellow, and homozygous derived alleles blue.
Exponentially fitted curves showing the decay of pairwise covariance for variable positions where Africans carry ancestral alleles and the Neandertal genome carries derived alleles.
A common model for the modern human colonization of Asia 23,28 assumes that an early coastal migration gave rise to the present-day people of Oceania, while a later more northern migration gave rise to Europeans and mainland Asians. The fact that the 45,000-year-old individual from Siberia is not more closely related to the Onge from the Andaman Islands (putative descendants of an early coastal migration) than he is to present-day East Asians, or Native Americans (putative descendants of a northern migration) (Fig. 3), shows that at least one other group to which the ancestors of the Ust’-Ishim individual belonged colonized Asia prior to 45,000 years ago. Interestingly, the Ust’-Ishim individual probably lived during a warm period (Greenland Interstadial 12) that has been proposed to be a time of expansion of modern humans into Europe 29,30. However, the latter hypothesis is based only on the appearance of the so-called “Initial Upper Paleolithic” industries (SI 5), and not on the identification of modern human remains 31,32. It is possible that the Ust’-Ishim individual was associated with the Asian variant of Initial Upper Paleolithic industry, documented at sites such as Kara-Bom in the Altai Mountains at about 47,000 yrs BP. This would then represent an early modern human radiation into Europe and Central Asia that may have failed to leave descendants among present-day populations29.
Online Methods
All sequencing was performed on the Illumina HiSeq 2000 and base-calling was carried out using Ibis 1.1.6 33. Reads were merged and remaining adaptor sequences trimmed before being aligned to the Human reference genome (GRCh37/1000 Genomes) using BWA (version 0.5.10) 34. GATK version 1.3 (v1.3–14-g348f2b) was used to produce genotype calls for each site. We excluded from analysis tandem repeats and regions of the genome that are not unique. We considered only genomic regions that fall within the 95% coverage distribution (SI 7) and where at least 99 % of overlapping 35mers covering a position map uniquely, allowing 1 mismatch.
Acknowledgments
We are grateful to Philipp Gunz, Martin Kircher, Andrey I. Krivoshapkin, Philip Nigst, Matthias Ongyerth, Nick Patterson, Gabriel Renaud, Udo Stenzel Mark Stoneking and Sahra Talamo for valuable input, comments and help; Theresa Pfisterer and Heiko Temming for technical assistance. Q.F. is funded in part by Chinese Academy of Sciences (XDA05130202) and the Ministry of Science and Technology of China (2007FY110200); P.A.K. by Urals Branch, Russian Academy of Sciences (12-C-4-1014) and Y.V.K. by the Russian Foundation for Basic Sciences (12-06-00045); F.J. and M.S. by the National Institutes of Health of the USA (R01-GM40282); and P.J. by NIH (K99-GM104158). D.R. is a Howard Hughes Medical Institute Investigator and supported by National Science Foundation (1032255) and the NIH (GM100233). Major funding for this work was provided by the Presidential Innovation Fund of the Max Planck Society.
Footnotes
All sequence data have been submitted to the European Nucleotide Archive (ENA) and are available under the following accession: Ust’-Ishim: PRJEB6622. The data from the 25 present-day human genomes are available as a public dataset from http://aws.amazon.com/datasets/ and from http://cdna.eva.mpg.de/neandertal/altai/.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
Contributions
Q.F., S.M.S., A.A.B., Y.V.K., J.K., B.V. and S.P. designed the research. A.P. and Q.F. performed the experiments; Q.F., H.L., P.M., F.J., P.L.F.J., K.P., C.d.F., M.M., M.L., M.K., D.R., J.K. and S.P. analysed genetic data; K.D. & T.F.G.H. performed 14C dating; D.C.S.G. & M.P.R. analyzed stable isotope data; N.V.P., P.A.K. & D.I.R. contributed samples and data; S.M.S., A.A.B., N.Z., Y.V.K., S.G.K., J.J.H. and B.V. analyzed archaeological and anthropological data; Q.F., J.K., B.V. and S.P. wrote and edited the manuscript with input from all authors.
 1,2
1,2